Home » Projects » Rangeland Soil Management and Hydrology » Watershed-scale Hydropedology » Hydrologic Modeling in Oak Woodland Soilscapes » SFREC » Elevation Survey by RTK » Establishing the Optimal Number of Survey Points » Selection by Minimizing MAE (mean absolute error)
Selection by Minimizing MAE (mean absolute error)
- 5 meter interval sampling of contours
- 5 meter interval sampling of terrain skeleton
- [5 10 15 20 25 30 40 50 60] meter thinning (snapping) radius
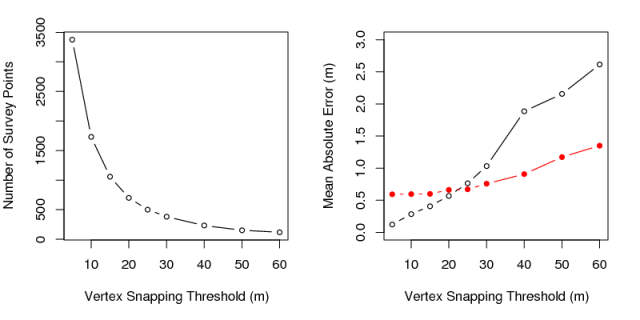
MAE vs. Thinning radius: terrain skeleton approach in red
# # prep work:
g.region res=10
v.to.rast in=w_buff out=w_buff_mask use=val val=1 --o
# invoked like this
# time sh eval_pts_spacing.sh 2>/dev/null
# approx 7 mins
# start logfie
outfile='stats.out'
echo '' > $outfile
# the min distance between new points when converting lines to points
# make this a small number for lots of points
dmax=5
# sequence of thinning thresholds
sequence="5 10 15 20 25 30 40 50 60"
# loop over final cleaning snapping threhold
for j in $sequence
do
# feedback
echo "thinning at : $j meters"
# setup some kind of sampling intervals
skeleton_sampling_interval=$j
# create a set of points along contours
v.to.points --q -i -t in=contours out=cp_00001 dmax=$dmax --o
v.category --q in=cp_00001 out=cp_cat_00001 option=del --o
v.category --q in=cp_cat_00001 out=cp_cat_1_00001 option=add --o
# select only those within the watershed:
v.select --q --o -t ainput=cp_cat_1_00001 atype=point binput=w_buff btype=area operator=overlap out=contour_pts_00001
# convert to points on a regular interval
# use a small number to preserve the complexity--
# we will thin the number of points down later
v.to.points --q -i -t in=skel out=sp_00001 dmax=$dmax --o
# these points need a category first
#
v.category --q in=sp_00001 out=sp_cat_00001 option=del --o
v.category --q in=sp_cat_00001 out=sp_cat_1_00001 option=add --o
# only points within watershed
v.select -t --o --q ainput=sp_cat_1_00001 atype=point binput=w_buff btype=area operator=overlap out=skel_pts_00001
# give each sampling point a unique ID and attr designating the observation type:
v.db.addtable --q --o skel_pts_00001 2>/dev/null
v.db.addtable --q --o contour_pts_00001 2>/dev/null
v.db.addcol --q --o skel_pts_00001 column='elev double'
v.db.addcol --q --o contour_pts_00001 column='elev double'
# patch contour and skeleton sampling points
v.patch --o --q -e in=contour_pts_00001,skel_pts_00001 out=rp_00001
v.build --q rp_00001
# thin points out a bit
v.clean --o --q in=rp_00001 out=rtk_pts_00001 tool=snap thres=$skeleton_sampling_interval
# extract number of points:
# save to $points
eval `v.info rtk_pts_00001 -t | grep points`
echo "$points points along terrain skeleton"
#extract elevation at each sampling points
v.what.rast --q vect=rtk_pts_00001 raster=e10 column=elev
# interpolate elevation for current region
# at the same time capture the number of points used in the interpolation
interp_points=`v.surf.rst in=rtk_pts_00001 zcol=elev elev=e_interp_00001 maskmap=w_buff_mask --o | grep 'The number of points from vector map is' | awk -F'is ' '{print $2}'`
# generate n random points, where n= number of points used in terrain skeleton-based interpolation
# note that these are generated along the grid used for sampling
# only generate points within watershed mask
r.random --o in=e10 n=$interp_points vector=rand_00001 cover=w_buff_mask
# interpolate elevation for current region (random points)
v.surf.rst in=rand_00001 zcol=value elev=rand_interp_00001 maskmap=w_buff_mask --o
# compute MAE
r.mapcalc "e_diff_00001 = abs(e10 - e_interp_00001)"
mae=`r.univar e_diff_00001 | grep 'mean:' | awk -F': ' '{print $2}'`
# compute MAE
r.mapcalc "rand_diff_00001 = abs(e10 - rand_interp_00001)"
rand_mae=`r.univar rand_diff_00001 | grep 'mean:' | awk -F': ' '{print $2}'`
# save stats
echo "$j,$points,$interp_points,$mae,$rand_mae" >> $outfile
done| thinning | points | interp_points | mae | rand_mae | |
|---|---|---|---|---|---|
| 1 | 5 | 3375 | 3318 | 0.59 | 0.12 |
| 2 | 10 | 1730 | 1693 | 0.60 | 0.28 |
| 3 | 15 | 1058 | 1040 | 0.60 | 0.41 |
| 4 | 20 | 698 | 691 | 0.66 | 0.57 |
| 5 | 25 | 501 | 495 | 0.67 | 0.76 |
| 6 | 30 | 381 | 379 | 0.76 | 1.03 |
| 7 | 40 | 231 | 227 | 0.91 | 1.89 |
| 8 | 50 | 152 | 150 | 1.17 | 2.15 |
| 9 | 60 | 117 | 117 | 1.35 | 2.61 |
Projects
- BMP's for Irrigated Agriculture
- Pedology and Soil Survey
- Rangeland Soil Management and Hydrology
- Watershed-scale Hydropedology
- Hydrologic Modeling in Oak Woodland Soilscapes
- Watershed-scale Hydropedology